Food industrial processing can influence its chemistry and composition, thus studying such an impact is relevant to provide new knowledge regarding their safety and possible advantageous effects on quality parameters.
Historically, food science has had a reductionist approach, focusing on the analysis of single compounds derived from food. However, new technological advances are making it possible to analyse simultaneously hundreds or thousands of compounds typically occurring in food products with one method by using their specific chemical properties in mass spectrometry analysis.
These food-derived compounds are known as metabolites and are present in cells, tissues, organs, and organisms. They include amino acids, sugars, fatty acids, or vitamins. The simultaneous analysis and characterisation of these metabolites is commonly termed metabolomic profiling or fingerprinting, and the methodology itself is called “Metabolomics.”
Why study the metabolites?
Studying the metabolites can provide a snapshot of what is going on in an organism, for example in fruits, vegetables, or more complex food, as a result of different food treatments. In this context, the methodology is also called “foodomics”. It allows for an understanding of the impact of external factors such as temperature, stress, microorganisms, environmental contaminants, toxins, and industrial processing on foods.
Therefore, on the one hand, foodomics is a valuable and promising tool for food processors and scientists to use in determining changes in the metabolome of food, including its biochemistry and composition. On the other hand, it can provide a benefit to the final consumer as food bioactivity, safety, quality, and traceability can be studied, possibly leading to better products.
How do we study the metabolome?
Studying the biochemical complexity in food through metabolomics requires state-of-the-art instruments and bioinformatic tools. A combination of chromatography and mass spectrometry is used to obtain spectral signals that are converted into numerical matrices, which allow for calculating potential differences between samples.
The number of variables generated can be considered as “BigData”, and complex statistics are required for the interpretation of the data, often with the aid of graphs and plots. In-house and public metabolite libraries are studied in the attempt to identify the compounds that are different between samples with the aim to find a biological interpretation of the observed changes.
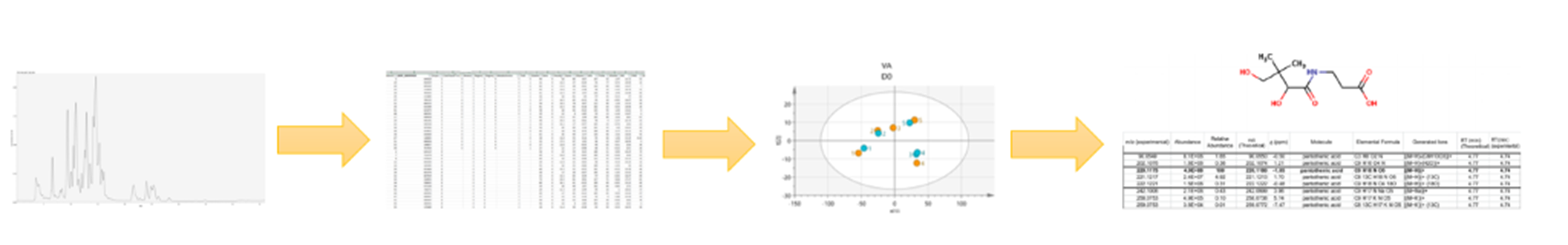
Data processing in metabolomics, from mass spectra to numerical data, including statistical analysis and metabolite identification. Illustration: Oscar Daniel Rangel Huerta
(Et avsnitt om metabolomikkforskningen i iNOBox oppdateres og settes inn her før lansering).
Test english
Test content English fact box.

